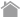MR1 Tetramer
The reagent to detect MAIT cells
What are MAIT cells?
 Mucosa-associated invariant T (MAIT) cells are classified as innate T cells, and are considered to constitute the largest T cell subpopulation in the human body. Initially, these cells were named after their localization in the lamina propria of the intestinal mucosa. However, it has been reported that they are frequently detected in the liver and among peripheral blood mononuclear cells (PBMCs).
Mucosa-associated invariant T (MAIT) cells are classified as innate T cells, and are considered to constitute the largest T cell subpopulation in the human body. Initially, these cells were named after their localization in the lamina propria of the intestinal mucosa. However, it has been reported that they are frequently detected in the liver and among peripheral blood mononuclear cells (PBMCs).
MAIT cells are thought to respond to bacterial and fungal infections by recognizing microbial vitamin B metabolites presented by MR1 molecules. MAIT cells are activated through engagement of these metabolites with the T cell receptor (TCR), leading to production of IFN-γ, IL-17, IL-2, and granzyme B. Thus, MAIT cells exert cytotoxic activities on infected cells, and prevent the spread of an infection.
What is MR1?
The MHC class I-related protein MR1 is a membrane protein non-covalently bound to β2-microglobulin (β2m), which is known as one of the non-classical MHC class I molecules. It is expressed in almost all cell types of the body. Under steady-state conditions, it is not present on the cell surface, but localized in the endoplasmic reticulum. Upon stimulation, for example, during an infection, MR1 molecules associated with microbial vitamin B metabolites migrate to the cell surface to present them to and thereby activate MAIT cells.
What is an MR1 Tetramer?
An MR1 Tetramer is a reagent prepared by tetramerizing biotinylated human MR1/β2m complexes with the help of phycobiliprotein-labeled streptavidin.
Human diseases related to MAIT cells
MAIT cells have been proposed to act as innate T cells that primarily respond to bacterial and fungal antigens. Recently, MAIT cells were found to be associated with autoimmune diseases. In addition, it has been reported that intestinal bacteria play a role in the development and differentiation of MAIT cells, therefore gaining increasing attention by researchers in the field of intestinal immunity.
Systemic lupus erythematosus, rheumatoid arthritis, ankylosing spondylitis,
multiple sclerosis, type I diabetes, fibromyalgia syndrome,
inflammatory bowel disease; ulcerative colitis, Crohn's disease
Common ligands for MR1
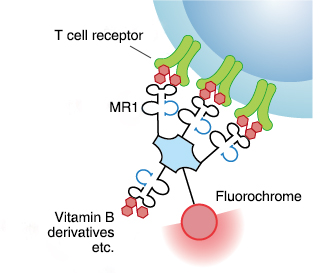
| Name | rRL-6-CH2OH: reduced 6-hydroxymethyl- 8-D-ribityllumazine | 5-OP-RU: 5-(2-oxopropylideneamino)- 6-D-ribitylaminouracil | 6-FP: 6-formylpterin |
|---|---|---|---|
| Feature | The first ligand identified for MR12); MR1 loaded with this ligand can activate MAIT cells |
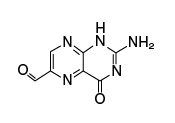
This ligand has high affinity for MR1; it is synthesized from intermediates of the early stages of vitamin B2 biosynthesis |
Photodegradation product of folic acid; MR1-6-FP complex cannot activate MAIT cells |
| Structure | 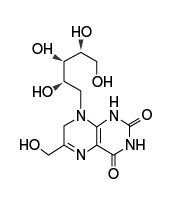 |
 |
 |
How to prepare ligand-loaded MR1 Tetramer?
*The product does NOT include the MR1 ligand.
5-OP-RU
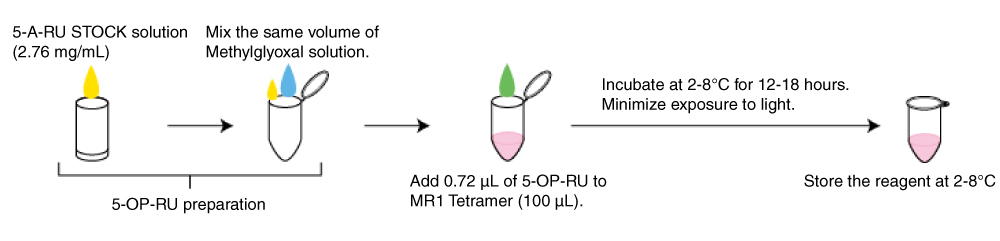
5-A-RU STOCK solution: 5-A-RU (Toronto Research Chemicals, Code No. A629245) 2.76 mg/mL (in ddw)
Methylglyoxal solution: Mix 1 µL of methylglyoxal (Sigma, M0252-25ML) and 406 µL of water.
Add 0.72 µL of 5-OP-RU to MR1 Tetramer (100 µL).
Ac-6-FP [A ligand for negative control of MR1 tetramer]
Ac-6-FP (Carbosynth, Code No. FA16935) 2 mg/mL (in 10 mM NaOH)
Add 1 µL of Ac-6-FP to 10 µL of MR1 Tetramer.
Example 1: Cell staining using PE-labeled MR1 Tetramers
PBMCs from healthy donors were collected from freshly isolated heparinized peripheral blood according to standard methods. These PBMCs were stained with T-Select Human MR1 Tetramer v2-PE loaded with 5-OP-RU or Ac-6-FP, as described in the datasheet.
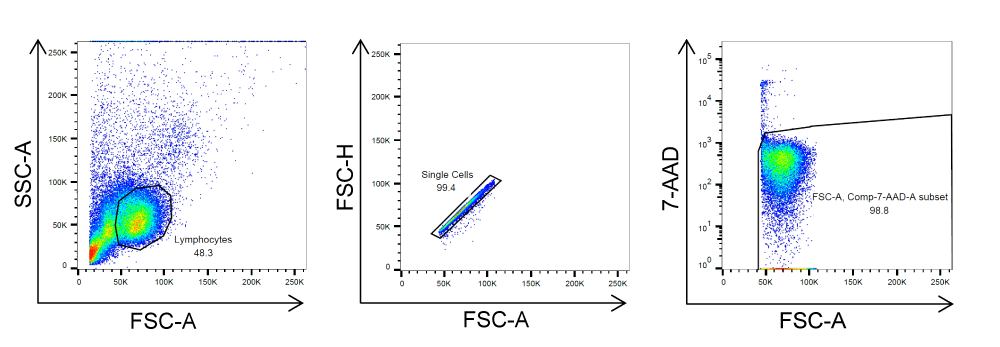
The single lymphocyte population was defined using the FSC/SSC and FSC-H/FSC-A gates, and the viable cell population was defined using the FSC/7-AAD gate.
Gating
Result
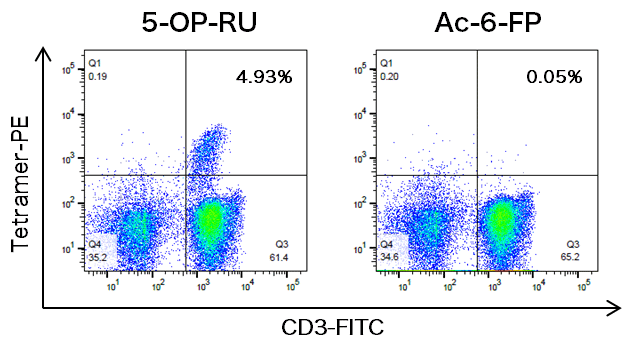
*Numbers in the upper right quadrants represent the percentages of T-select Human MR1 Tetramer+ cells relative to the total CD3+ cells.

Please check the product information. [Click here!]
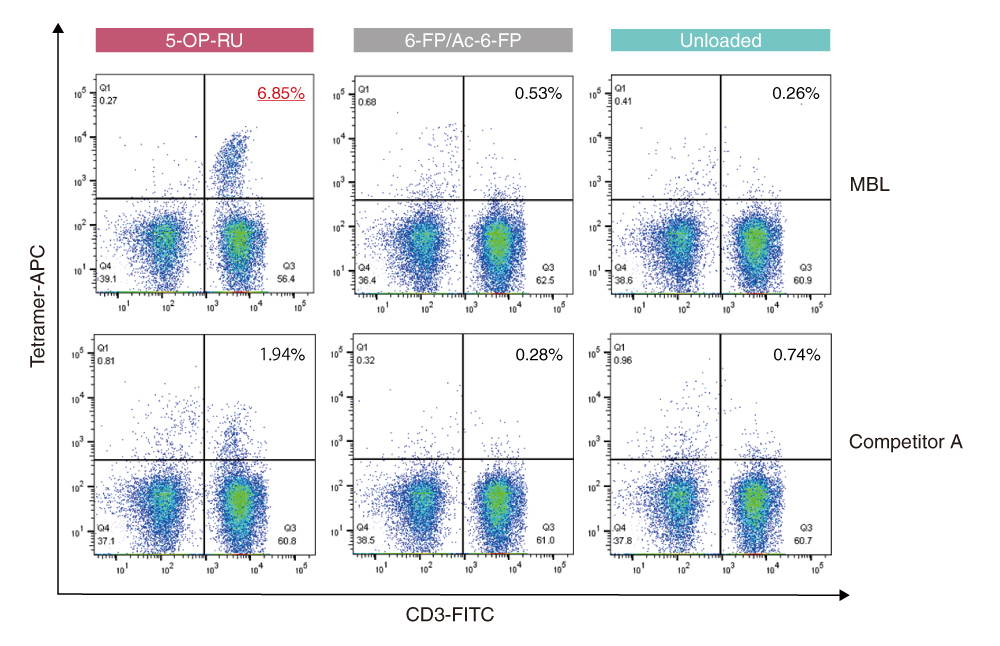
Example 2: Staining comparison of MR1 Tetramer products
PBMCs from healthy donors were stained with T-Select Human MR1 Tetramer v2-APC loaded with 5-OP-RU or Ac-6-FP, or unloaded. It was compared with staining data using human MR1 tetramer of competitor A loaded with 5-OP-RU or 6-FP, or unloaded.
The single lymphocyte population was defined using the FSC/SSC and FSC-H/FSC-A gates, and the viable cell population was defined using the FSC/7-AAD gate.
References
1) Dusseaux M, et al. Blood 117:1250-1259 (2011)
2) Kjer-nielsen L, et al. Nature 491:717-723 (2012)
3) Reantragoon R, et al. J Exp Med 210:2305-2320 (2013)
4) Corbett AJ, et al. Nature 509:361-365 (2014)
5) Eckle SB, et al. J Exp Med 211:1585-1600 (2014)
6) Howson LJ, et al. Front Immunol 6:303 (2015)
7) Mondot S, et al. Immunogenetics 68:537-548 (2016)
8) Keller AN, et al. Nat Immunol 18:402-411 (2017)
9) Keller AN, et al. Curr Opin Immunol 46:66-74 (2017)
10) Greene JM, et al. Mucosal Immunol 10:802-813 (2017)
11) Kjer-nielsen L, et al. Immunol Cell Biol 6:573-587 (2018)
12) Chiba A, et al. Front Immunol 9:1333 (2018)