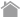Poster presentation at The 22nd Annual Meeting of the Japanese Society for Chronobiology
Analysis of mRNA and Protein Expression Profiles of Core Circadian Genes
Shun Matsuzawa, Hitomi Nakamura , Yoshiko Takashima , Noriko Ishikawa , Yoshiro Kishi
Medical & Biological Laboratories Co., Ltd. (MBL)
Abstract
The expression of circadian genes has been assessed mainly by quantitating their mRNAs using qRT-PCR. However, to understand the molecular mechanisms of circadian rhythm, it is essential to quantitate the amount of proteins that actually perform the genes’ functions and analyze the regulation of proteins by post-translational protein modifications such as phosphorylation and ubiquitination. In fact, post-translational modification of circadian clock proteins in the core loop includes phosphorylation and ubiquitination of PER and ubiquitination of CRY.
We have successfully generated specific antibodies to factors in the circadian core loop (Per1 and 2, Cry1 and 2, and Chrono). Here we report the time course of expression of these proteins in relation to the expression of their corresponding mRNAs.
Methods
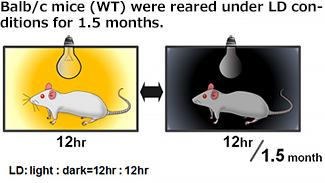
| Rearing | Sampling |
|---|---|
|
|
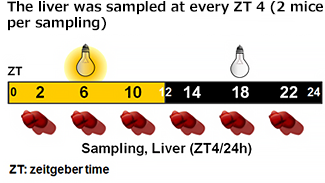
| Gene expression analysis |
|---|
* Relative mRNA levels are normalized to Gapdh mRNA. qRT-PCR data are means ± SEM (n=2). |
Results
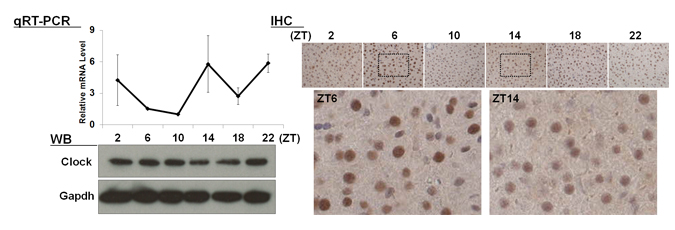
| 【 Clock 】 Antibody used:Anti-CLOCK (Mouse) mAb (Code No. D349-3) |
|---|
|
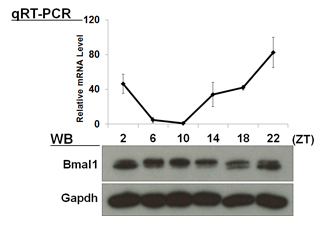
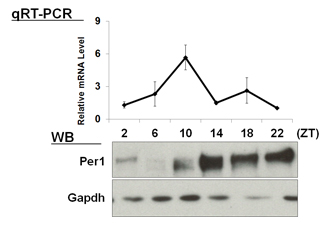
| 【 Bmal1 】 Antibody used:Anti-BMAL1 (Mouse) mAb (Code No. D335-3) | 【 Per1 】 Antibody used:Anti-Per1 (Mouse) pAb (Code No. PM091) |
|---|---|
|
|
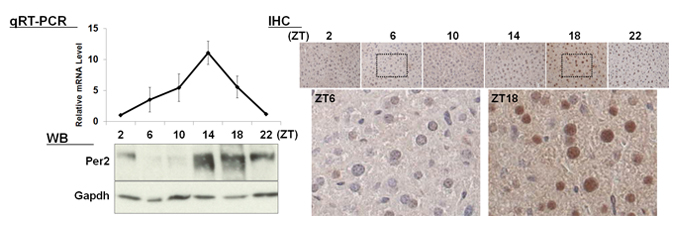
| 【 Per2 】 Antibody used:Anti-Per2 (Mouse) pAb (Code No. PM083) |
|---|
|
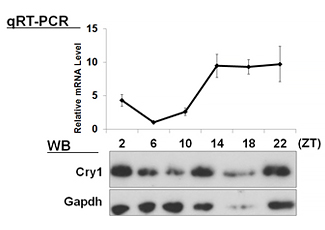
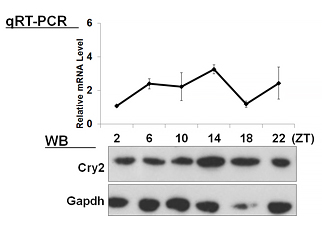
| 【 Cry1 】 Antibody used:Anti-Cry1 (Mouse) pAb (Code No. PM081) | 【 Cry2 】 Antibody used:Anti-Cry2 (Mouse) pAb (Code No. PM082) |
|---|---|
|
|
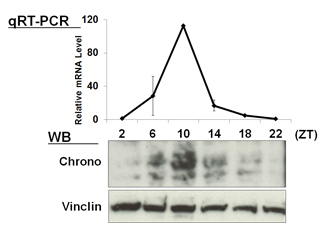
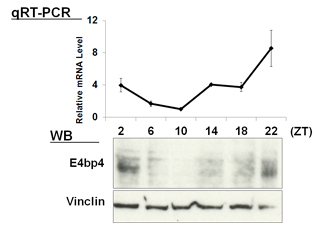
| 【 Chrono (Gm129) 】 Antibody used:Anti-Chrono (Mouse) pAb (Code No. PM087) | 【 E4bp4 (Nfil3) 】 Antibody used:Anti-NFIL3 (E4BP4) chimeric mAb (Code No. M225-3) |
|---|---|
|
|
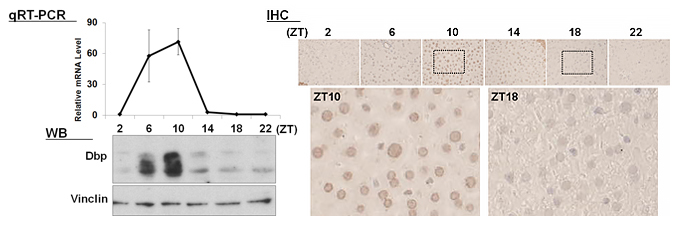
| 【 Dbp 】 Antibody used:Anti-DBP (Mouse) pAb (Code No. PM079) |
|---|
|
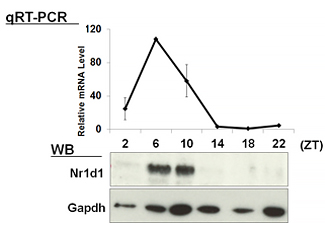
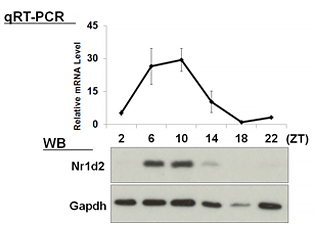
| 【 Nr1d1 (Rev-erb α) 】 Antibody used:Anti-NR1D1 (Rev-erbα) pAb (Code No. PM092) | 【 Nr1d2 (Rev-erb β) 】 Antibody used:Anti-NR1D2 (Rev-erbβ) pAb (Code No. PM093) |
|---|---|
|
|
Conclusion
The importance of studying protein expression is suggested.
Not only did the amount of proteins change over time, but also the banding pattern due to post-translational modification of the proteins (Bmal1, Per1, Per2, E4bp4, and Dbp). These changes stem from the regulation of gene expression that cannot be monitored by measuring changes in the amount of mRNA, strongly suggesting the importance of monitoring protein expression specifically.
Acknowledgement
Primer sequences for Gapdh, Cry2, Bmal1, Clock, Per1, Per2, Dbp, E4bp4, Rev-erb α, and Rev-erb β were provided by Dr. Hiroki Ueda (RIKEN Quantitative Biology Center).